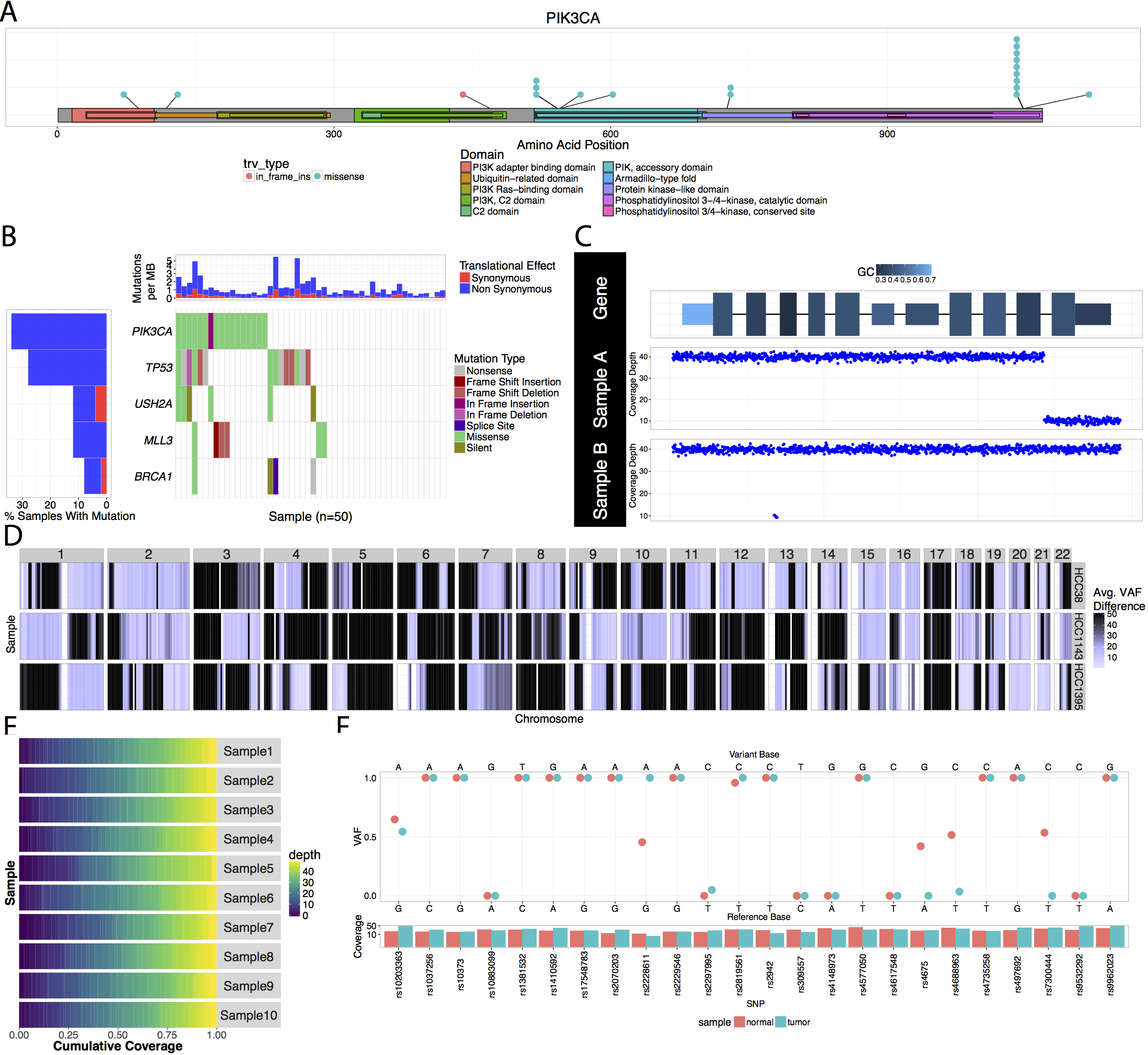
Genomic Data Visualization and Interpretation
The age of affordable massively parallel sequencing has exponentially increased the availability of genome, transcriptome, epigenome, and other kinds of omic profiling data. Extracting biologically meaningful results and communicating this information remains challenging. This site aims to expose users to a number of “best-in-class” tools and methods for overcoming this challenge.
We aim to provide an in-depth tutorial in the interpretation and visualization of omic data. Among the topics we will cover are: “Genome Browsing and Interpretation”, “Differential Expression and Pathway Analysis”, “Best in Class tools for visualization”, and others. Students should have a background in biology and a basic knowledge of the R programming language and linux.
This resource is structured in a modular format. Users would benefit from familiarity with topics covered in previous modules however it is not a strict requirement unless otherwise stated. Please navigate to Course to begin.
These materials have been developed with the support of Physalia-courses and Evomics-workshops. The first in-person workshop was held at Freie Universität Berlin, Germany, 11-15 September 2017. The second iteration was an abbreviated one-day version included as part of the Evomics Workshop on Genomics in Cesky Krumlov, Czech Republic, 6-19 January 2019. The third iteration of this course, with improved and updated content, was held at Freie Universität Berlin, Germany, 8-12 April 2019 (see course page). For future offerings by Physalia (including this workshop) please visit the upcoming workshops page. For future offerings by Evomics please visit Evomics workshops.
Physalia-courses provides scientific training courses and workshops in bioinformatics, genomics and related fields, promoting the transfer of new methods and emerging techniques to a broad range of researchers. Their goal is to build a knowledge-sharing platform between highly qualified instructors and participants at various stage of their scientific and academic career. Through the courses and workshops, participants learn how to plan their projects and how to analyze their data.
Evomics (Evolution and Genomics) is currently organizing three Workshops, the Workshop on Molecular Evolution, the Workshop on Genomics, and the Workshop on Population and Speciation Genomics. All three Workshops are designed to provide advanced training, much of which is hands-on, in their respective fields. Workshops feature faculty who are demonstrated leaders in their fields and a dedicated team of instructors and technical staff to ensure that each Workshop is an efficient and effective learning experience for all participants.
We are constantly improving the course material, if you notice a problem or have a suggestion for improvement please let us know by making a github issue.
